shinybody is an htmlwidget of the human
body that allows you to hide/show and assign colors to 79 different body
parts. The human widget is an htmlwidget, so
it works in Quarto documents, R Markdown documents, or anything other
HTML medium. It also functions as an input/output widget in a
shiny app.
You can install the development version of shinybody
from GitHub with:
# install.packages("devtools")
devtools::install_github("robert-norberg/shinybody")You can install from CRAN with:
install.packages("shinybody")Here is a simple example of using the human widget in an
R Markdown document:
library(shinybody)
example_organs <- c("brain", "eye", "heart", "stomach", "bladder")
my_organ_df <- subset(shinybody::shinybody_organs, organ %in% example_organs)
my_organ_df$show <- TRUE
my_organ_df$color <- grDevices::rainbow(nrow(my_organ_df))
my_organ_df$selected[1] <- TRUE
my_organ_df$hovertext <- mapply(
function(o, clr) htmltools::strong(tools::toTitleCase(o), style = paste("color:", clr)),
my_organ_df$organ,
my_organ_df$color,
SIMPLIFY = FALSE
)
human(gender = "female", organ_df = my_organ_df)
Here is a complete list of the organs that are available:
#> Male Female
#> adipose_tissue ✅ ✅
#> adrenal_gland ✅ ✅
#> amygdala ✅ ✅
#> aorta ✅ ✅
#> appendix ✅ ✅
#> atrial_appendage ✅ ✅
#> bladder ✅ ✅
#> bone ✅ ✅
#> bone_marrow ✅ ✅
#> brain ✅ ✅
#> breast ✅ ✅
#> bronchus ✅ ✅
#> caecum ✅ ✅
#> cartilage ✅ ✅
#> cerebellar_hemisphere ✅ ✅
#> cerebellum ✅ ✅
#> cerebral_cortex ✅ ✅
#> circulatory_system ✅ ✅
#> colon ✅ ✅
#> coronary_artery ✅ ✅
#> diaphragm ✅ ✅
#> duodenum ✅ ✅
#> ectocervix ❌ ✅
#> endometrium ❌ ✅
#> epididymis ✅ ❌
#> esophagus ✅ ✅
#> eye ✅ ✅
#> fallopian_tube ❌ ✅
#> frontal_cortex ✅ ✅
#> gall_bladder ✅ ✅
#> gastroesophageal_junction ✅ ✅
#> heart ✅ ✅
#> ileum ✅ ✅
#> kidney ✅ ✅
#> left_atrium ✅ ✅
#> left_ventricle ✅ ✅
#> liver ✅ ✅
#> lung ✅ ✅
#> lymph_node ✅ ✅
#> mitral_valve ✅ ✅
#> nasal_pharynx ✅ ✅
#> nasal_septum ✅ ✅
#> nerve ✅ ✅
#> nose ✅ ✅
#> oral_cavity ✅ ✅
#> ovary ❌ ✅
#> pancreas ✅ ✅
#> parotid_gland ✅ ✅
#> penis ✅ ❌
#> pituitary_gland ✅ ✅
#> placenta ❌ ✅
#> pleura ✅ ✅
#> prefrontal_cortex ✅ ✅
#> prostate_gland ✅ ❌
#> pulmonary_valve ✅ ✅
#> rectum ✅ ✅
#> renal_cortex ✅ ✅
#> salivary_gland ✅ ✅
#> seminal_vesicle ✅ ❌
#> skeletal_muscle ✅ ✅
#> skin ✅ ✅
#> small_intestine ✅ ✅
#> smooth_muscle ✅ ✅
#> spinal_cord ✅ ✅
#> spleen ✅ ✅
#> stomach ✅ ✅
#> submandibular_gland ✅ ✅
#> temporal_lobe ✅ ✅
#> testis ✅ ❌
#> throat ✅ ✅
#> thyroid_gland ✅ ✅
#> tongue ✅ ✅
#> tonsil ✅ ✅
#> trachea ✅ ✅
#> tricuspid_valve ✅ ✅
#> uterine_cervix ❌ ✅
#> uterus ❌ ✅
#> vagina ❌ ✅
#> vas_deferens ✅ ❌Here is a very simple shiny app using the human
widget:
library(shiny)
library(shinybody)
male_organs <- shinybody_organs$organ[shinybody_organs$male]
female_organs <- shinybody_organs$organ[shinybody_organs$female]
ui <- function() {
fluidPage(
selectInput(
inputId = "gender",
label = "Select Gender",
choices = c("male", "female"),
multiple = FALSE,
selected = "male"
),
selectInput(
inputId = "body_parts",
label = "Select Body Parts to Show",
choices = male_organs,
multiple = TRUE,
selected = male_organs[1:5]
),
humanOutput(outputId = "human_widget"),
verbatimTextOutput(outputId = "clicked_body_part_msg"),
verbatimTextOutput(outputId = "selected_body_parts_msg")
)
}
server <- function(input, output, session) {
observe({
g <- input$gender
if (g == "male") {
organ_choices <- male_organs
} else {
organ_choices <- female_organs
}
updateSelectInput(
session = session,
inputId = "body_parts",
choices = organ_choices,
selected = organ_choices[1:5]
)
})
output$human_widget <- renderHuman({
selected_organ_df <- subset(shinybody::shinybody_organs, organ %in% input$body_parts)
selected_organ_df$show <- TRUE
human(
gender = input$gender,
organ_df = selected_organ_df,
select_color = "red"
)
})
output$clicked_body_part_msg <- renderPrint({
paste("You Clicked:", input$clicked_body_part)
})
output$selected_body_parts_msg <- renderPrint({
paste("Selected:", paste(input$selected_body_parts, collapse = ", "))
})
}
shinyApp(ui = ui, server = server)
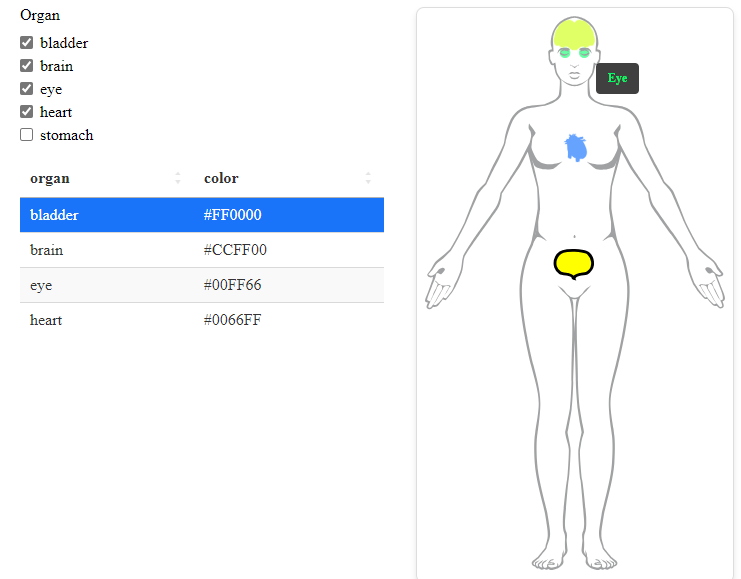
shinybody is crosstalk compatible. Here is
an example of a simple crosstalk widget using
shinybody and DT.
library(shinybody)
library(DT)
example_organs <- c("brain", "eye", "heart", "stomach", "bladder")
my_organ_df <- subset(shinybody::shinybody_organs, organ %in% example_organs)
my_organ_df$show <- TRUE
my_organ_df$color <- grDevices::rainbow(nrow(my_organ_df))
my_organ_df$selected[1] <- TRUE
my_organ_df$hovertext <- mapply(
function(o, clr) htmltools::strong(tools::toTitleCase(o), style = paste("color:", clr)),
my_organ_df$organ,
my_organ_df$color,
SIMPLIFY = FALSE
)
my_organ_df_shared_data <- crosstalk::SharedData$new(my_organ_df)
checkboxes <- crosstalk::filter_checkbox(
id = "filter",
label = "Organ",
sharedData = my_organ_df_shared_data,
group = ~organ
)
tbl <- DT::datatable(
data = my_organ_df_shared_data,
options = list(
pageLength = 10,
columnDefs = list(
list(visible = FALSE, targets = c("male", "female", "show", "selected", "hovertext"))
)
),
rownames = FALSE,
height = "500px",
autoHideNavigation = TRUE
)
crosstalk::bscols(
htmltools::tagList(checkboxes, tbl),
human(gender = "female", organ_df = my_organ_df_shared_data),
device = "sm"
)